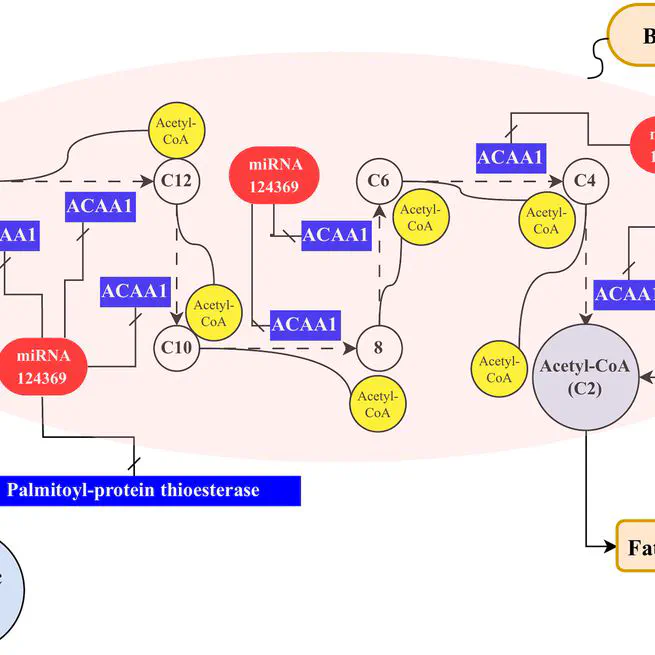
MicroRNAs (miRNAs) are key regulators of gene expression in plants, influencing various biological processes such as oil quality and seed development. Although, our knowledge about miRNAs in olive (Olea europaea L.) is progressing, with several miRNAs being identified in previous studies, but most of these reported miRNAs have been predicted without the aid of a reference genome, primarily due to limited genome accessibility at the time. However, significant knowledge gaps still need to be improved in this area. This study addresses the complexities of miRNA detection in olive, using a high quality reference genome and a combination of genomics and machine learning-based methods. By leveraging random forest and support vector machine algorithms, we successfully identified 56 novel miRNAs in olive, surpassing the limitations of conventional homology-based methods. Our subsequent analysis revealed that some of these miRNAs are implicated in the regulation of key genes involved in oil quality. Within the context of oil biosynthesis pathways, the novel miRNA Oeu124369 regulates fatty acid biosynthesis by targeting acetyl-CoA acyltransferase 1 and palmitoyl-protein thioesterase, thereby influencing the production of acetyl-CoA and palmitic acid, respectively. These findings underscore the power of machine learning in unraveling the complex miRNA regulatory network in olive and provide a high quality miRNA resource for future research aimed at improving olive oil production by exploring the target genes of the identified miRNAs to understand their role and their biological processes.
Oct 11, 2024
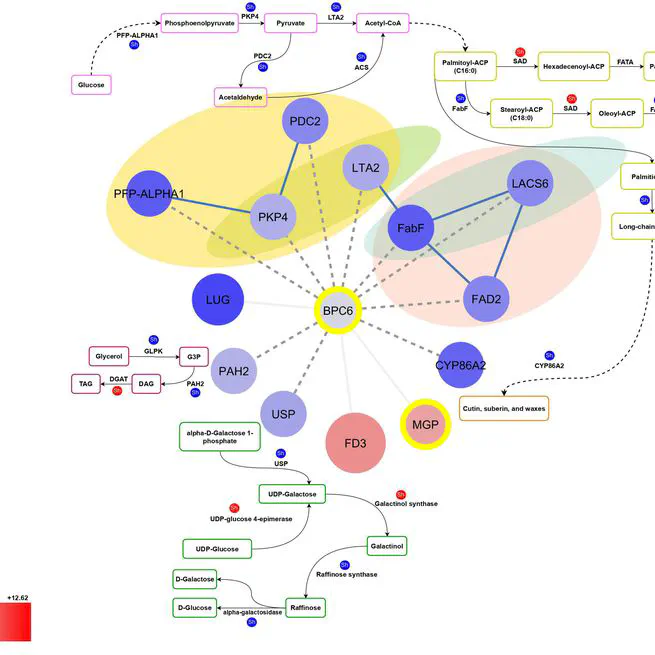
The balanced fatty acid profile of olive oil not only enhances its stability but also contributes to its positive effects on health, making it a valuable dietary choice. Olive oil's high content of unsaturated fatty acids and low content of saturated fatty acids contribute to its beneficial effects on cardiovascular diseases and cancer. The quantities of these fatty acids in olive oil may fluctuate due to various factors, with genotype being a crucial determinant of the oil's quality. This study investigated the genetic basis of oil quality by comparing the transcriptome of two Iranian cultivars with contrasting oil profiles; Mari, known for its high oleic acid content, and Shengeh, characterized by high linoleic acid at Jaén index four. Gas chromatography confirmed a significant difference in fatty acid composition between the two cultivars. Mari exhibited significantly higher oleic acid content (78.48%) compared to Shengeh (48.05%), while linoleic acid content was significantly lower in Mari (4.76%) than in Shengeh (26.69%). Using RNA sequencing at Jaén index four, we analyzed genes involved in fatty acid biosynthesis. Differential expression analysis identified 2775 genes showing statistically significant differences between the cultivars. Investigating these genes across nine fundamental pathways involved in oil quality led to the identification of 25 effective genes. Further analysis revealed 78 transcription factors and 95 transcription binding sites involved in oil quality, with BPC6 and RGA emerging as unique factors. This research provides a comprehensive understanding of the genetic and molecular mechanisms underlying oil quality in olive cultivars. The findings have practical implications for olive breeders and producers, potentially streamlining cultivar selection processes and contributing to the production of high-quality olive oil.
Oct 1, 2024

A deep search of RNA-seq published data shed light on thirty-nine experiments associated with the olive transcriptome, four of these proved to be ideal for meta-analysis. Meta-analysis confirmed the genes identified in previous studies and released new genes, which were not identified before. According to the IDR index, the meta-analysis had good power to identify new differentially expressed genes. The key genes were investigated in the metabolic pathways and were grouped into four classes based on the biosynthetic cycle of fatty acids and factors that affect oil quality. Galactose metabolism, glycolysis pathway, pyruvate metabolism, fatty acid biosynthesis, glycerolipid metabolism, and terpenoid backbone biosynthesis were the main pathways in olive oil quality. In galactose metabolism, raffinose is a suitable source of carbon along with other available sources for carbon in fruit development. The results showed that the biosynthesis of acetyl-CoA in glycolysis and pyruvate metabolism is a stable pathway to begin the biosynthesis of fatty acids. Key genes in oleic acid production as an indicator of oil quality and critical genes that played an important role in production of triacylglycerols were identified in different developmental stages. In the minor compound, the terpenoid backbone biosynthesis was investigated and important enzymes were identified as an interconnected network that produces important precursors for the synthesis of a monoterpene, diterpene, triterpene, tetraterpene, and sesquiterpene biosynthesis.
Aug 22, 2023
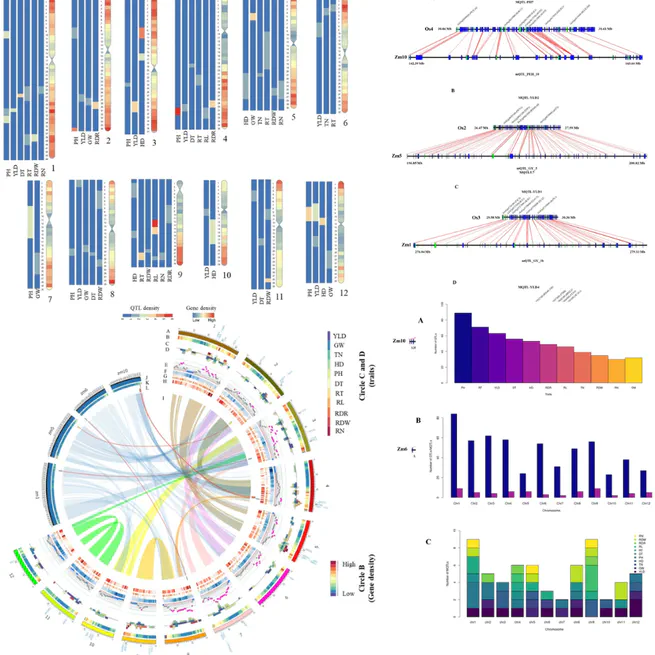
Meta-QTL (MQTL) analysis is a robust approach for genetic dissection of complex quantitative traits. Rice varieties adapted to non-flooded cultivation are highly desirable in breeding programs due to the water deficit global problem. In order to identify stable QTLs for major agronomic traits under water deficit conditions, we performed a comprehensive MQTL analysis on 563 QTLs from 67 rice populations published from 2001 to 2019. Yield and yield-related traits including grain weight, heading date, plant height, tiller number as well as root architecture-related traits including root dry weight, root length, root number, root thickness, the ratio of deep rooting and plant water content under water deficit condition were investigated. A total of 61 stable MQTLs over different genetic backgrounds and environments were identified. The average confidence interval of MQTLs was considerably refined compared to the initial QTLs, resulted in the identification of some well-known functionally characterized genes and several putative novel CGs for investigated traits. Ortho-MQTL mining based on genomic collinearity between rice and maize allowed identification of five ortho-MQTLs between these two cereals. The results can help breeders to improve yield under water deficit conditions.
Mar 25, 2021