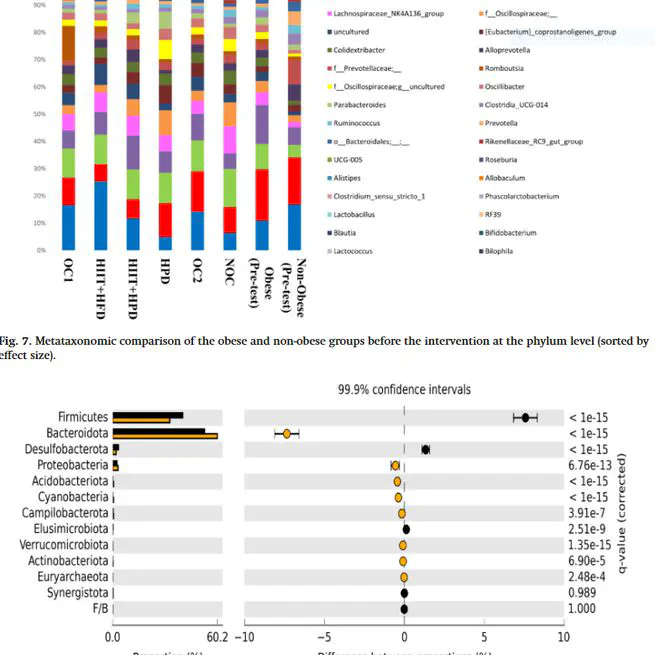
Diet and exercise are two critical factors that regulate gut microbiota, affecting weight management. The present study investigated the effect of 10 weeks of high-intensity interval training (HIIT) and a high-protein diet (HPD) on gut microbiota composition and body weight changes in obese male Wistar rats. Forty obese rats were randomly divided into five groups, including HPD, HIIT + HPD, HIIT + high-fat diet (HFD) (continuing HFD during intervention), obese control 1 (continuing HFD during intervention), obese control 2 (cutting off HFD at the beginning of the intervention and continuing standard diet), and eight non-obese Wistar rats as a non-obese control (NOC) group (standard diet). Microbial community composition and diversity analysis by sequencing 16S rRNA genes derived from the fecal samples, body weight, and Lee index were assessed. The body weight and Lee index in the NOC, HIIT + HFD, HPD, and HIIT + HPD groups were significantly lower than that in the OC1 and OC2 groups along with the lower body weight and Lee index in the HPD and HIIT + HPD groups compared with the HIIT + HFD group. Also, HFD consumption and switching from HFD to a standard diet or HPD increased gut microbiota dysbiosis. Furthermore, HIIT along with HFD increased the adverse effects of HFD on gut microbiota, while the HIIT + HPD increased microbial richness, improved gut microbiota dysbiosis, and changed rats’ phenotype to lean. It appears that HFD discontinuation without doing HIIT does not improve gut microbiota dysbiosis. Also, the HIIT + HFD, HPD, and HIIT + HPD slow down HFD-induced weight gain, but HIIT + HPD is a more reliable strategy for weight management due to its beneficial effects on gut microbiota composition.
Aug 29, 2023
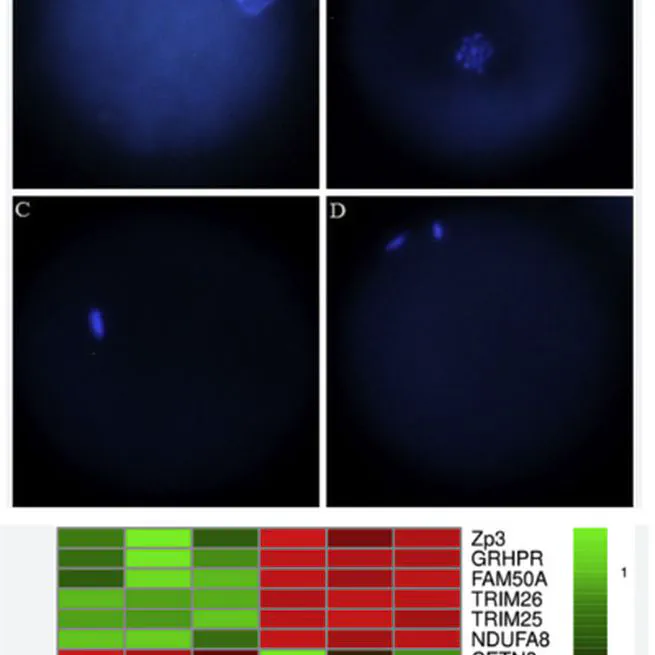
Lipopolysaccharide (LPS) derived from gram negative bacteria cell wall is known to cause ruminal acidosis and/or infectious diseases such as metritis and mastitis which has a significant negative impact on the reproductive performance. This study aimed to investigate the effect of LPS on oocyte maturation and subsequent development in vitro. Ovine cumulus oocyte complexes (COCs) were matured in a medium supplemented with 0 (control), 0.01, 0.1, 1 and 10 μg/mL LPS. Nuclear maturation, cleavage and blastocyst rate, mitochondrial membrane potential (ΔΨm), intracellular reactive oxygen species (ROS) content and changes to the transcript abundance were evaluated. In case of the maturation rate, the percentage of oocytes reaching the MII stage was lower following exposure to 10 μg/mL LPS in comparison to the control group (P < 0.05). Moreover, the blastocyst rate decreased in case of 1 and 10 μg/mL LPS when compared to the control group (P < 0.05). ROS overproduction accompanied by a decreased ΔΨm were recorded in LPS treated oocytes in comparison to the control group (P < 0.05). The 3′ tag digital gene expression profiling method revealed that 7887 genes were expressed while only seven genes exhibited changes in the transcript abundance following exposure to LPS. Tripartite motif containing 25 (TRIM25), Tripartite motif containing 26 (TRIM26), Zona Pellucida glycoprotein 3 (ZP3), Family with sequence similarity 50-member A (FAM50A), Glyoxalate and hydroxy pyruvate reductase (GRHPR), NADH ubiquinase oxireductase subunit A8 (NDUFA8) were down-regulated (P < 0.05), while only Centrin 3 (CETN3) was up-regulated (P < 0.05). Our findings show that LPS has undesirable effects on the maturation competence of ovine oocytes and subsequent embryo development. In addition, the transcriptomic profiling results may shed more light on the molecular mechanisms of LPS-induced infertility in ruminants.
Nov 1, 2020
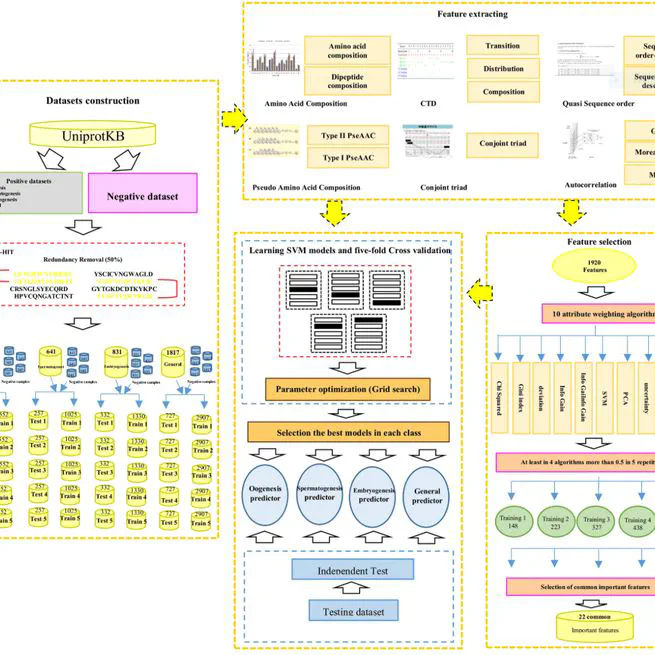
Successful spermatogenesis and oogenesis are the two genetically independent processes preceding embryo development. To date, several fertility-related proteins have been described in mammalian species. Nevertheless, further studies are required to discover more proteins associated with the development of germ cells and embryogenesis in order to shed more light on the processes. This work builds on our previous software (OOgenesis_Pred), mainly focusing on algorithms beyond what was previously done, in particular new fertility-related proteins and their classes (embryogenesis, spermatogenesis and oogenesis) based on the support vector machine according to the concept of Chou’s pseudo-amino acid composition features. The results of five-fold cross validation, as well as the independent test demonstrated that this method is capable of predicting the fertility-related proteins and their classes with accuracy of more than 80%. Moreover, by using feature selection methods, important properties of fertility-related proteins were identified that allowed for their accurate classification. Based on the proposed method, a two-layer classifier software, named as “PrESOgenesis” (https://github.com/mrb20045/PrESOgenesis) was developed. The tool identified a query sequence (protein or transcript) as fertility or non-fertility-related protein at the first layer and then classified the predicted fertility-related protein into different classes of embryogenesis, spermatogenesis or oogenesis at the second layer.
Jun 13, 2018