A comprehensive transcriptomic meta-Analysis leveraging deep learning to uncover molecular signatures and potential therapeutic targets in Triple-Negative Breast Cancers
Apr 9, 2025··
0 min read
S Salesi, T Adel, Z Nayeri, v Shariati, a Zomorodipour
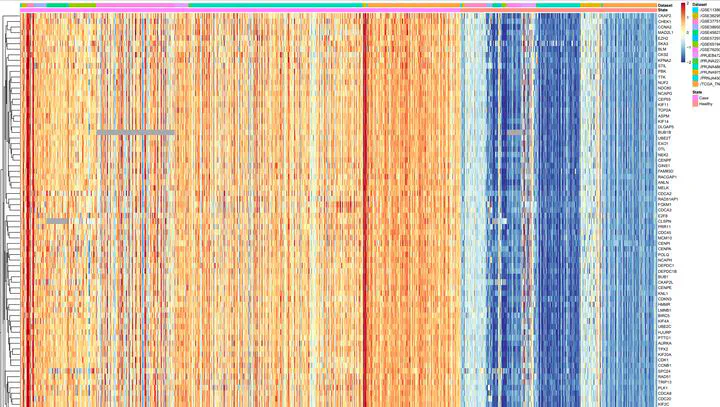 Figure 8. Clustring of metagenes
Figure 8. Clustring of metagenesAbstract
Triple-negative breast cancer (TNBC), characterized by its aggressive behavior and lack of hormone receptor expression, remains a therapeutic challenge. This study integrates multi-omics data and AI-driven approaches to dissect the molecular mechanisms driving TNBC progression. Through a meta-analysis of 49 transcriptomic studies (2013–2024), we identified 2,101 differentially expressed genes (DEGs), including 68 consistently dysregulated protein-coding genes, with CXCL10 (↑4.01-fold) and ADH1B (↓4.8-fold) as the most significantly altered. Pathway enrichment revealed upregulated genes associated with cell proliferation, immune evasion, and metabolic reprogramming, while downregulated genes implicated hormonal signaling suppression and extracellular matrix remodeling. Gene Ontology analysis highlighted mitotic regulation and immune dysregulation as central processes. AI-based clustering of protein-protein interaction networks identified five functional modules (Tumor Growth, Invasion & Metastasis, Metabolism, Immune & Inflammation, Hormonal & Stress Response), with hub genes like CDK1 and CXCL8 driving tumor proliferation and immune escape. Notably, machine learning algorithms enhanced data integration and cluster identification, revealing FOXM1 as a key regulator of mitotic pathways (p = 6.189E-07) and JUN as a mediator of stromal-epithelial interactions despite its downregulation. Hormonal profiling uncovered systemic suppression of estrogen-responsive (ESR1, FOXA1) and neurohormonal (ADRB1, AGTR1) pathways, emphasizing TNBC’s endocrine-silenced phenotype. Immune analysis demonstrated dual chemokine dysregulation, loss of homeostatic signals (CXCL12, CCL21) impaired immune surveillance, while pro-inflammatory chemokines (CXCL8, CCL20) recruited immunosuppressive cells. Chronic interferon signaling (STAT1, ISG15) and immune checkpoint alterations (CTLA4, CD80/86) further shaped a resistant microenvironment. These findings elucidate TNBC’s complex molecular landscape, integrating AI-enhanced analytics to prioritize therapeutic targets, such as mitotic kinases, chemokine networks, and hormonal signaling nodes, offering actionable insights for overcoming therapy resistance in this aggressive subtype.
Type