
A deep search of RNA-seq published data shed light on thirty-nine experiments associated with the olive transcriptome, four of these proved to be ideal for meta-analysis. Meta-analysis confirmed the genes identified in previous studies and released new genes, which were not identified before. According to the IDR index, the meta-analysis had good power to identify new differentially expressed genes. The key genes were investigated in the metabolic pathways and were grouped into four classes based on the biosynthetic cycle of fatty acids and factors that affect oil quality. Galactose metabolism, glycolysis pathway, pyruvate metabolism, fatty acid biosynthesis, glycerolipid metabolism, and terpenoid backbone biosynthesis were the main pathways in olive oil quality. In galactose metabolism, raffinose is a suitable source of carbon along with other available sources for carbon in fruit development. The results showed that the biosynthesis of acetyl-CoA in glycolysis and pyruvate metabolism is a stable pathway to begin the biosynthesis of fatty acids. Key genes in oleic acid production as an indicator of oil quality and critical genes that played an important role in production of triacylglycerols were identified in different developmental stages. In the minor compound, the terpenoid backbone biosynthesis was investigated and important enzymes were identified as an interconnected network that produces important precursors for the synthesis of a monoterpene, diterpene, triterpene, tetraterpene, and sesquiterpene biosynthesis.
Aug 22, 2023
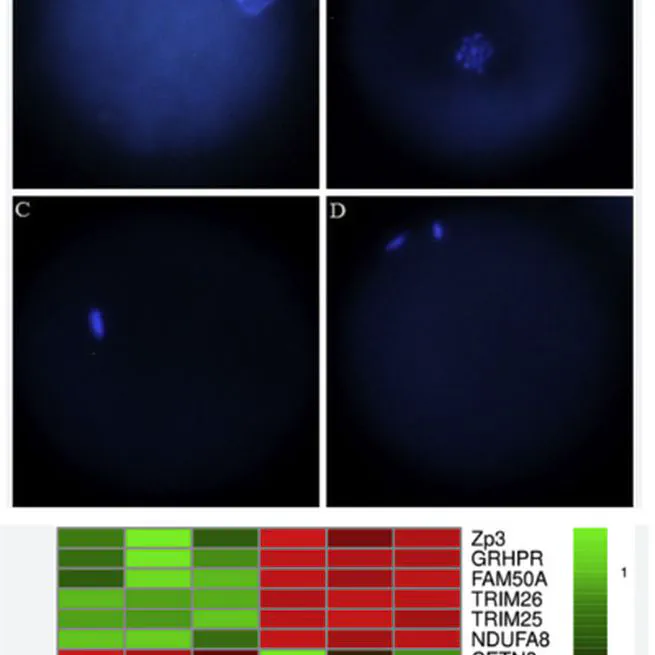
Lipopolysaccharide (LPS) derived from gram negative bacteria cell wall is known to cause ruminal acidosis and/or infectious diseases such as metritis and mastitis which has a significant negative impact on the reproductive performance. This study aimed to investigate the effect of LPS on oocyte maturation and subsequent development in vitro. Ovine cumulus oocyte complexes (COCs) were matured in a medium supplemented with 0 (control), 0.01, 0.1, 1 and 10 μg/mL LPS. Nuclear maturation, cleavage and blastocyst rate, mitochondrial membrane potential (ΔΨm), intracellular reactive oxygen species (ROS) content and changes to the transcript abundance were evaluated. In case of the maturation rate, the percentage of oocytes reaching the MII stage was lower following exposure to 10 μg/mL LPS in comparison to the control group (P < 0.05). Moreover, the blastocyst rate decreased in case of 1 and 10 μg/mL LPS when compared to the control group (P < 0.05). ROS overproduction accompanied by a decreased ΔΨm were recorded in LPS treated oocytes in comparison to the control group (P < 0.05). The 3′ tag digital gene expression profiling method revealed that 7887 genes were expressed while only seven genes exhibited changes in the transcript abundance following exposure to LPS. Tripartite motif containing 25 (TRIM25), Tripartite motif containing 26 (TRIM26), Zona Pellucida glycoprotein 3 (ZP3), Family with sequence similarity 50-member A (FAM50A), Glyoxalate and hydroxy pyruvate reductase (GRHPR), NADH ubiquinase oxireductase subunit A8 (NDUFA8) were down-regulated (P < 0.05), while only Centrin 3 (CETN3) was up-regulated (P < 0.05). Our findings show that LPS has undesirable effects on the maturation competence of ovine oocytes and subsequent embryo development. In addition, the transcriptomic profiling results may shed more light on the molecular mechanisms of LPS-induced infertility in ruminants.
Nov 1, 2020